Project Overview
Created by Olisemeka Nmarkwe and Sujana Mehta, this project implements a robust data mining pipeline for the analysis of EEG (electroencephalogram) signals. The pipeline extracts meaningful statistical features and discovers latent structure through unsupervised clustering. The workflow begins with rigorous signal preprocessing (including filtering and referencing), followed by automated statistical feature generation in windowed segments. These engineered features serve as the basis for KMeans and hierarchical clustering, with results assessed via a suite of quantitative metrics and visual analytics. The approach highlights practical ML engineering skills: transforming complex biomedical data into actionable insights and building modular, scalable analytic code.
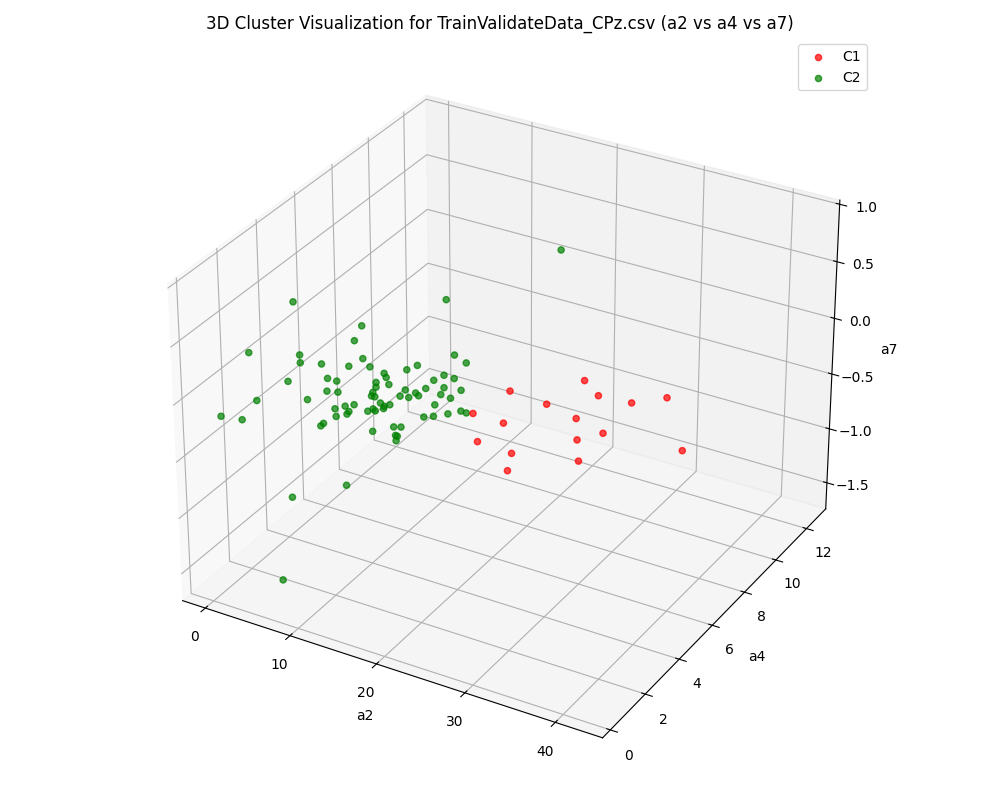
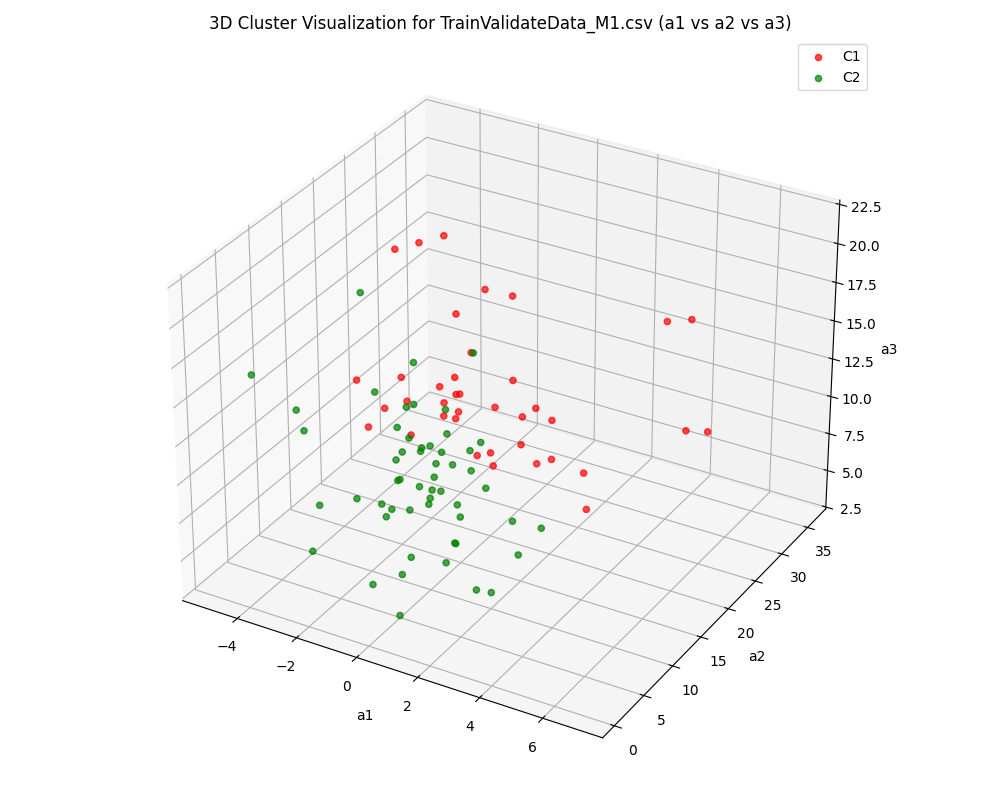
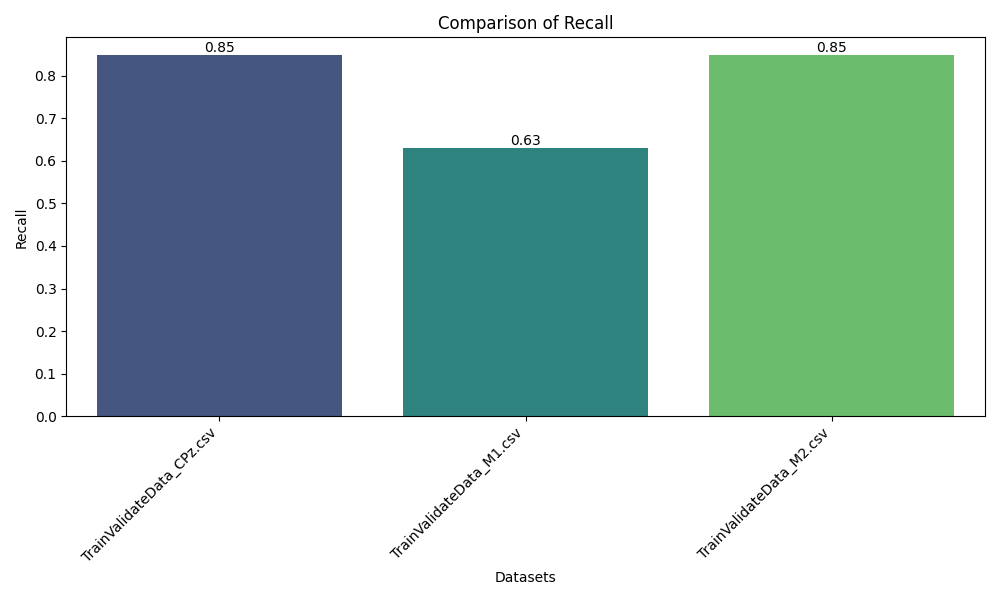
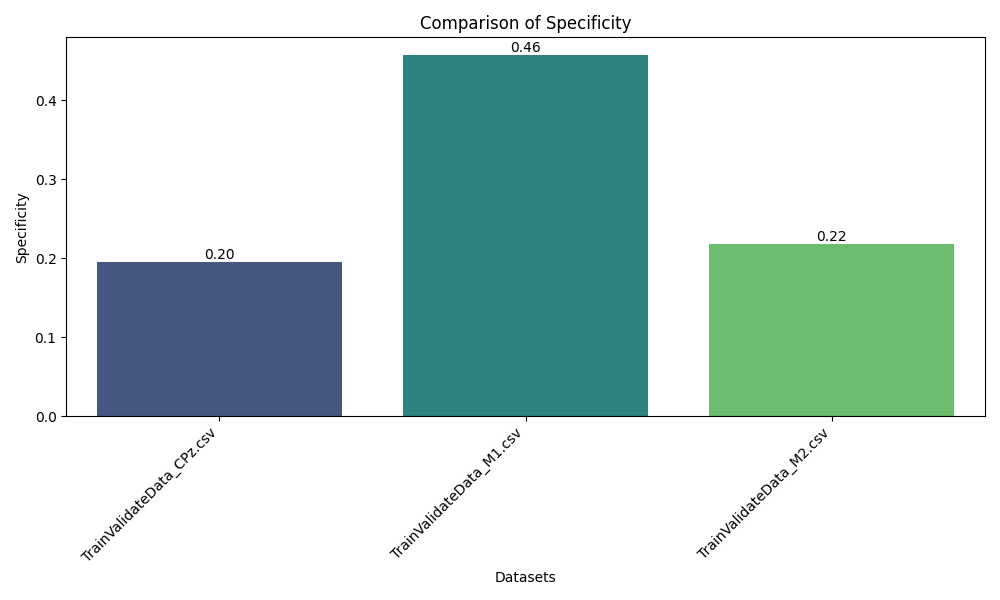
A key aspect of the project is its comparative analysis across three distinct EEG channels—CPz (central parietal), M1, and M2 (mastoid references)—demonstrating how different brain signal locations impact clustering outcomes and the separability of neural states.
Key Features
- EEG Data Preprocessing: Cleans and structures raw multichannel EEG data using bandpass, notch, and impedance filtering; handles re-referencing to improve signal quality.
- Automated Feature Extraction: Generates windowed statistical features (mean, standard deviation, kurtosis, skewness) to summarize signal characteristics for machine learning.
- Flexible Data Pipeline: Modular design allows easy extension to new subjects, sessions, or EEG channels.
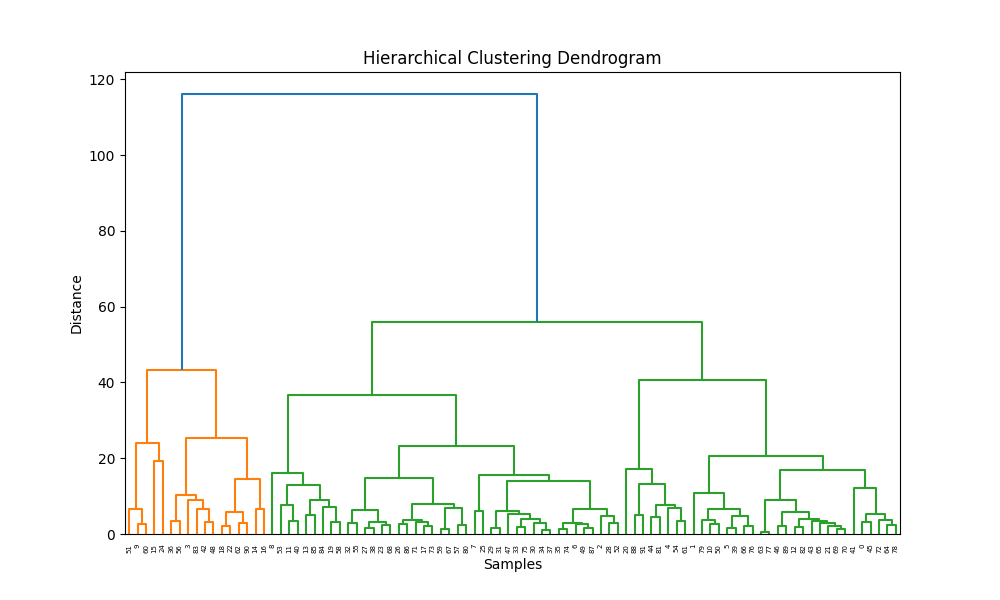
- Unsupervised Clustering: Applies KMeans and Agglomerative (hierarchical) clustering to engineered features; supports PCA-based dimensionality reduction for visualization and model input.
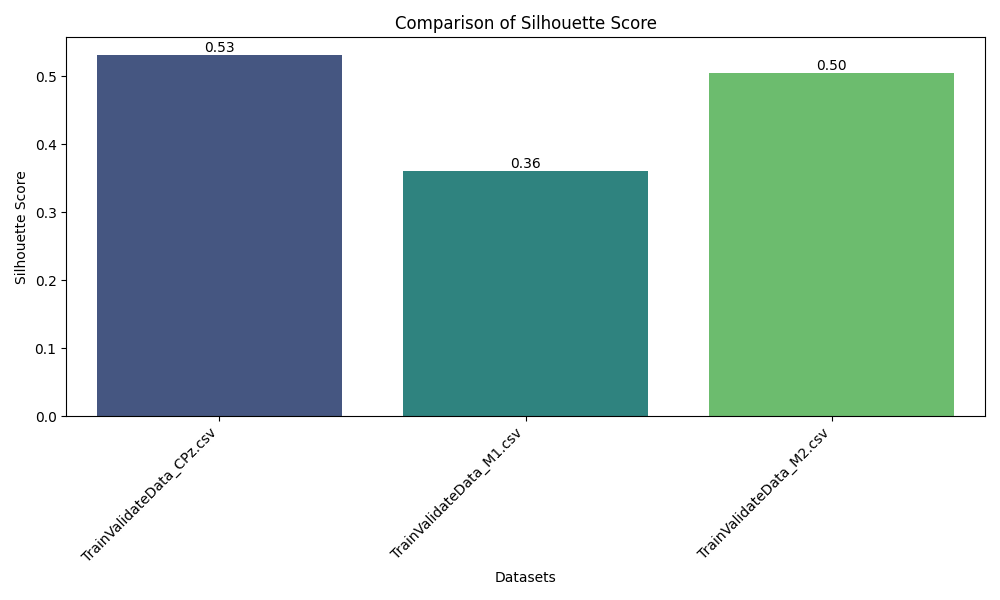
- Metric-Driven Evaluation: Evaluates clustering outcomes using Silhouette Score, Cohesion, Separation, Recall, Specificity, and F1-Score—bridging unsupervised and supervised evaluation principles.
- Comprehensive Visualization: Automates generation of distribution plots, bar charts, and cluster visualizations, supporting interpretability and reporting.
- Reproducible Outputs: Saves preprocessed datasets, feature matrices, and plots for further ML tasks or validation.
- Multi-Channel Comparison: Examines and compares clustering results for CPz, M1, and M2 channels to understand their relative discriminative power for EEG-based analysis.
Technologies Used
- Python (core language for all modules)
- NumPy, Pandas (data manipulation, analytics)
- SciPy (signal processing, statistics)
- scikit-learn (clustering, metrics, preprocessing, PCA)
- Matplotlib, Seaborn (visualization and plotting)
- Pickle (data serialization)
- OS (file and directory management)
Results
The pipeline successfully transformed raw EEG recordings into a high-quality, feature-rich dataset suitable for machine learning. Clustering analysis revealed distinct groupings corresponding to underlying brain states, validated by high Silhouette and separation scores across subjects and sessions. Comparative analysis across CPz, M1, and M2 channels illustrated the superior discriminative power of the CPz channel for neural state differentiation. The modular pipeline supports rapid reconfiguration for new datasets or evaluation strategies, and the visualization suite aids both diagnostic review and scientific presentation.
Future Work
Next steps include extending to semi-supervised and deep learning models, integrating domain-specific feature extraction (e.g., frequency bands), and exploring real-time applications for BCI (brain-computer interface) or cognitive state monitoring. The pipeline also provides a foundation for downstream supervised ML tasks, such as classification or anomaly detection, using the extracted EEG features.
Screenshots